Bengt Hubendick was a genius in the way Picasso was a genius. It’s not that he didn't make mistakes. They just didn't matter, because that wasn't the point.
At left [1] is a “mean photograph” of Lymnaea peregra from
Hubendick’s 1951 monograph. Here he has
photographed 20 shells from a single population in Sweden, overlain the
negatives, and developed a single print.
Hubendick’s point here is variance.
Variance is the stuff of evolution.
And what has become of our science today? In April [2] we touched briefly on the 2010
paper by Ana Correa and her colleagues, synthesizing 15 years of research on
molecular evolution in the Lymnaeidae [3].
Essentially every one of the 53 tips on Correa’s tree was labeled with a
different specific nomen. There was no estimate of intraspecific sequence
variance whatsoever.
Nor am I able to find any such estimate anywhere in the
entire heap of 21 papers on lymnaeid molecular phylogenetics stacked
like cordwood on my desk this afternoon [4].
All 21 seem to adhere strictly to what might be called the “U1s2nmt3 Rule.” Workers usually sequence one individual per
population, sometimes two, and never more than three. And they usually sequence one population per
nominal species, sometimes two, and never more than three. Apparently there has been some international
conference of bonehead tree jocks where a U1s2nmt3 Rule was proposed and
adopted without debate, to which I was not privy.
But gene trees are (at best) weak, null hypotheses of
population relationships [5]. They
require control and calibration, no different from any other measurement
technique in the toolbox of evolutionary biology. When applied to estimate the genetic relationships
among a set of populations unknown to us, we first need an estimate of the
variance within populations, and then between populations within species.
So in April I asked the rhetorical question, “Do we know
enough about the biology of the lymnaeid snails to make sense out of all the
sequence data published in recent years by Remigio, Bargues, Mas-Coma,
Pfenninger, Streit, Jarne, Pointier, and others?” The answer is, “Maybe we know enough about Lymnaea stagnalis.”
Back in the Glory Days of the Modern Synthesis, Hubendick
suggested that the lymnaeid fauna of North America might share as many as three
species with that of Europe. And much less problematic than L. palustris, about
which we have obsessed during the months of April and May, was Lymnaea
stagnalis.
Lymnaea stagnalis is such a striking snail that there has
apparently never been any question, from the birth of American malacology to
the present day, that the big honkin’ lymnaeids with the concave apex and the
flaring lip crawling around in our northern lakes must match the big honkin’
lymnaeids with the concave apex and the flaring lip crawling around in lakes
throughout northern Europe.
In the 20th Century L. stagnalis became a popular
laboratory model for neurobiologists, parasitologists, and physiologists of all
stripes, to the point that my simple search of the NCBI PubMed database (for
example) yielded 1,344 hits. Lymnaea
stagnalis may surpass Biomphalaria glabrata (1,222 hits) for the mantle of
“best known freshwater gastropod.”
So I went fishing in GenBank for L. stagnalis sequences in
an effort to calibrate the Correa gene tree.
My initial hope was to analyze the same set of genes used by Correa and
colleagues – 16S, ITS1 and ITS2 – but I found no ITS sequences for L. stagnalis
from the American side of the pond. The
eight sequences for 16S in GenBank (from four sources) included four Canadian
and four European, I think [6], but there were some irregularities in those
data, which gave me pause.
The best dataset in GenBank seemed to be that for CO1, in
any case, with four Canadian and eight European sequences from six different
sources, all over 600 bp in length, so that’s what I focused on. The figure above shows a neighbor-joining
tree based on simple percent sequence differences among four CO1 sequences from
Manitoba, six from Germany (I think) and one each from Sweden and France (Click for larger). The scale bar is a bit misleading, but
divergence within continents ranged up to about 5%, with divergence values
between continents in the 7-9% range.
That’s sort-of consistent with the 16S situation, but the
16S sequences were only in the 400-450 bp range, and the divergence didn’t look
nearly as clean as in the CO1 data I’ve figured. The GenBank accessions suggested that Correa
and colleagues may have obtained a 16S sequence in an L. stagnalis sampled from
France that matched one of Remigio’s Manitoba sequences at 100%. Meanwhile the maximum divergence across the
whole set of eight was between Remigio’s Canadian and Italian sequences, at
13%.
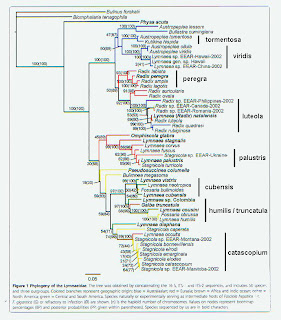
In any case, if we re-examine the Correa tree using 5%
expected sequence divergence within-continent [4] and 10% between, we obtain
the groupings I’ve labeled on the right margin in the figure below (Click for larger). From North America (that yellowish color in
Correa’s figure, hard to distinguish) we see one group comprising nominal
catascopium, emarginata, elodes and the “occulta” sample from Poland we
mentioned in April. We also note that
individual samples of caperata, megasoma, and columella were distinctive – no
surprises there. And there’s evidence
for a “Radix” population up in Canada matching Eurasian luteola – that’s
interesting.
The two other North American groups suggested by Correa’s
gene tree include the little mud-dwelling lymnaeids we here tend to call
“fossarines.” The situation with L. humilis and L. cubensis will, I think, bear closer examination. But I’m clean out of patience with this
subject for today, and I suspect the attention of my readership may be wearing
thin as well. So let’s sign off with
what should be a standard disclaimer for inquiries of this sort.
A species is a population or group of populations
reproductively isolated from all others.
Gene trees may or may not reflect reproductive isolation [7]. So the N = 8 groups (and N = 11 singletons) I
have marked on Correa’s gene tree are not “species,” even though I have labeled
them that way. There’re just gene
sequences. More about which next time…
Notes
[1] Plate 2, Figure 3 from Hubendick, B. (1951) "Recent Lymnaeidae, Their Variation,
Morphology, Taxonomy, Nomenclature and Distribution. Kungl. Svenska Vetenskapskademiens
Handlingar. Fjarde Serien Band 3. No. 1. Stockholm: Almquist & Wiksells
Boktryckeri AB.
[3] Correa, A. C., J. S. Escobar, P. Durand, F. Renaud, P.
David, P. Jarne, J-P Pointier, and S. Hurtrez-Bousses (2010) Bridging gaps in the molecular phylogeny of
the Lymnaeidae (Gastropoda: Pulmonata), vectors of fascioliasis. BMC Evolutionary Biology 10: 381. Open access here: [html]
[4] In fairness, Correa and colleagues (Infection, Genetics
& Evolution 11: 1978-1988) published a “DNA barcoding analysis” in 2011 similar
in spirit to the L. stagnalis study I have developed above. Their combined analysis of sequence
divergence across multiple studies and multiple species yielded a “barcoding gap”
(presumably marking the boundary between intraspecific and interspecific
variability) around 6%, quite consistent with my within-continent
estimate. Correa et al. (2011) did not
examine an intercontinental subset, however.
[6] Most of the sequences I inspected in GenBank were
missing locality data! A few authors supplied information regarding the site of
collection in the “source” field. But most
just entered “source = mitochondria.” Duh.
[7] What is a Species Tree? [12July11]