Editor’s Notes – This essay is the second half of a two part series on the systematics of the pleurocerid genus Leptoxis. We recommend that you familiarize yourself with last month’s post [6Apr23] before proceeding.
This essay was subsequently published as: Dillon, R.T., Jr. (2023b) Testing the Periwinkle Hypothesis. Pp 101 – 110 in The Freshwater Gastropods of North America Volume 6, Yankees at The Gap, and Other Essays. FWGNA Project, Charleston, SC.
I first met Nathan Whelan, that brash young star of find ‘em and grind ‘em malacology, at the Louisville meeting of the FMCS in April of 2011 [1]. He was bubbling with excitement about his dissertation research at the University of Alabama, “the first phylogenetic study of Leptoxis to have complete ingroup and adequate outgroup sampling.” Nathan’s 157-page dissertation, which included chapters on egg laying behavior and nuclear copies of mitochondrial genes (NUMTs), in addition to an extraordinarily ambitious molecular phylogeny, was published in 2013 [2].
Nathan analyzed N = 207 individual snails, focusing on 154 Leptoxis sampled from 39 populations of 13 nominal species, to which he added a very large number of outgroups: 23 Pleurocera/Elimia, 20 Lithasia, 4 Juga, an Io sample fished from Genbank, 2 Cleopatra from Zambia, and 3 “undescribed pleurocerids” from the Collins River for which he would not hazard even a genus.
He sequenced two nuclear genes (Histone H3 and 28S rRNA) and two mitochondrial genes (16sRNA and CO1) for his big sample, and immediately discovered 28 cases of mitochondrial superheterogeneity, recalling the phenomenon my coworkers and I reported in 2009 [3]. These highly divergent sequences he attributed to NUMTs, an hypothesis which his subsequent research did not confirm [4]. Soldiering on undaunted, he excluded 23 individual Leptoxis and 5 individual Lithasia from further analysis, paring his total sample size down to N = 179.
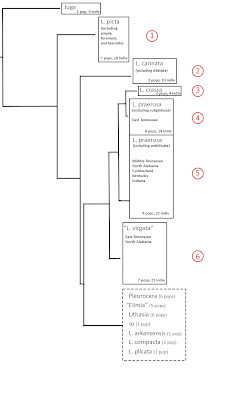 |
| Whelan [1] Fig 5.4, diagrammatic |
The figure above is a diagrammatic representation of Nathan’s Figure 5.4, a Baysian tree based on concatenation of the four genes, simplifying 179 branch tips [5] down to eight boxes. I would suggest that you right-click on that figure and open a larger version in a new window, because we're going to discuss it in some detail. And if you’d like to see Nathan's original tree, with all 179 tips unfolded and labelled, you are hereby invited to open [this link] in a third window as well [6]. And you'll need to max that third window up to life size. Nathan’s original Figure 5.4 was printed in 7-point font over two sequential 8.5 x 11 inch print pages, which I have pasted together into a single 15 x 20 inch jpeg image for detailed examination. OK, are we ready to go forward?
The box shown in dashed outline at the bottom of the diagrammatic representation symbolizes 26% of the foliage of the tree, a total of 47 sequences, including all the Pleurocera/Elimia, all the Lithasia, the Io and the unidentified. Mingled in amongst the branches bearing all those taxa are the sequences from three species of Leptoxis: L. compacta (5), L. plicata (2) and L. arkansensis (4). This is the phenomenon for which Nathan subsequently [7] coined the term, “prodigious polyphyly” implying that results such as these bear some relationship to the evolutionary relationships among the pleurocerid populations from which they were drawn.
But gene trees are dependent variables, not independent variables. Although they might certainly cast light on an hypothesis, they cannot be used to construct an hypothesis. And since I personally do not have enough research experience with Leptoxis compacta, L. plicata, or L. arkansensis to have formed an hypothesis regarding their evolutionary relationships [8], let’s trim the lower quarter of Nathan’s tree off, and set those data aside. One day we may be able to interpret them. But today is not that day.
Now moving forward with the 74% of Nathan’s tree for which, over 50 years of experience, I have developed an expectation. From top to bottom, six results present themselves for discussion.
First, Nathan’s results support the 1998 hypothesis of Dillon & Lydeard [9] that the Leptoxis of the Alabama/Coosa system are conspecific, Leptoxis picta (Conrad 1834) being the oldest available name. The nomina ampla (Anthony 1855), foremani (Lea 1843), and taeniata (Conrad 1834), are all junior synonyms of picta.
Second, Nathan’s results support the hypothesis of Dillon & Robinson [3] that populations previously identified as Leptoxis dilatata in the New/Greenbrier/Kanawha are trans-Appalachian Leptoxis carinata.
 |
| Leptoxis carinata eggs |
And third, Nathan’s results support the hypothesis of Dillon & Ahlstedt [10] that Leptoxis crassa is closely related to, but specifically distinct from, Leptoxis praerosa. Reproductive isolation seems to have arisen in an East Tennessee population of L. praerosa, rather than in Middle Tennessee, North Alabama, or elsewhere [11]. The subsequent level of divergence does not seem to be of the magnitude of a subgenus, much less a genus. The longstanding hypothesis that large-bodied, heavily shelled pleurocerid populations described as Melania crassa (Haldeman) or Melania anthonyi (Redfield) might be set aside in a unique genus “Eurycaelon” or “Athearnia” is not supported.
Fourth. Nathan’s gene tree depicts 8 populations of Leptoxis praerosa sampled from East Tennessee (four Clinch, one Powell, two Holston, one Nolichucky, comprising 14 individuals in total) as distinct from 9 populations (22 individuals) sampled from elsewhere throughout the remainder of the range. Nathan suggested that this phenomenon might reflect speciation, going so far as to nominate subglobosa (Say 1825) as a possible name for the East Tennessee populations.
There is no evidence of sympatry between the eight L. praerosa populations in Block 4 and the other nine populations in Block 5, however. Such a phenomenon is just as easily explained by geographic isolation as by reproductive isolation. Walden Ridge, constituting the eastern escarpment of the Cumberland Plateau as it slashes diagonally through Tennessee to Chattanooga, has long been recognized as an important biogeographic boundary separating the aquatic faunas of East and Middle Tennessee, and cannot be ruled out as a causative agent for the genetic discontinuity apparent between Block 4 and Block 5 above.
Fifth. Subsumed among the nine populations in the main Leptoxis praerosa Block 5 were two populations Nathan identified as Leptoxis umbilicata – one from the East Fork Stones River (5 individuals) and a second from Smith Fork of the Caney/Collins (3 individuals). There is no evidence of any genetic divergence between these two populations and Leptoxis praerosa sampled more broadly.
Indeed, our field surveys conducted throughout the Tennessee/Cumberland catchment have confirmed populations of otherwise typical L. praerosa bearing openly-umbilicate shells in the Stones River subdrainage as well as in the Red River subdrainage (NW of Nashville) and (sporadically) in the Elk. Intrapopulation variation such as that depicted from a tributary of the Red River below, together with the genetic data of Whelan, suggest that L. umbilicata (Wetherby 1876) be lowered to subspecific status [12] under Leptoxis praerosa.
 |
| Leptoxis praerosa umbilicata |
And sixth, and most interestingly. Nathan identified a set of 7 populations (21 individuals) from all around East Tennessee and North Alabama, in the Holston, Hiwassee, Nolichucky, Sequatchie, Paint Rock and elsewhere, as “Leptoxis virgata.” That subset of 7 populations seems to bear a distinct mitochondrial haplotype, differing by roughly 0.8 mystery units of divergence [13] from his 8 + 9 = 17 populations of L. praerosa. These 7 “virgata” populations seem effectively sympatric with praerosa populations over their entire range, collectable side-by-side in the Nolichucky River at the SR340 bridge, Greene County, TN.
The molecular distinction between L. praerosa and “L. virgata” seems to be entirely mitochondrial. Although Nathan included nuclear 28S and H3 data in his concatenated Fig 5.4 tree, the separate trees he figured for the two nuclear genes individually did not seem to resolve L. "virgata" from the greater Leptoxis background. The praerosa/”virgata” distinction does, however, seem to include a morphological component. Nathan stated that “the shells of these species are slightly different” but did not elaborate. You can be the judge from his Figure 4.5 below.
Nathan’s mitochondrial data plus the demonstrated sympatry plus the (putative) shell morphological differences all combine to suggest that a pair of cryptic Leptoxis species co-occur in the rivers of East Tennessee. And since the range of the more widespread (17-population) species extends north and west to include the Falls of The Ohio, the best identification for the 17-population species must be Leptoxis praerosa. Nathan’s identification of the 7-population species as “Leptoxis virgata” is, however, premature.
Before signing off on his dissertation, Nathan’s advisor should have sent him back to the Nolichucky River at the SR340 bridge to do the job right. Nathan should have been instructed to collect a big, fresh sample of Leptoxis, paying close attention to microhabitat, photograph them all, and measure their shells thoroughly. Then he should have sequenced them, and sorted them into two piles according to mtDNA haplotype. And then he should have cracked open a can of geometric morphometrics, or maybe an old-fashioned cigar box of discriminant function analysis, or maybe an even older-fashioned bivariate statistical analysis, to see if any difference in the morphology of those two piles of Leptoxis shells could be confirmed.
 |
| Whelan [2] Fig 4.5, modified |
In the Pleuroceridae, species are defined by shell morphology. So if Nathan had indeed been able to detect some shell morphological difference correlated with the mtDNA sequence divergence at the SR340 bridge, he should then have gone back into the national collections and conducted a similar morphological analysis on all the type specimens of all the Leptoxis available, or (in any case) their images as reproduced in their descriptions, and tried to find a match.
If, however, no shell morphological difference is detectable at the SR340 bridge, the pair of species may be judged true siblings. Then again, Leptoxis praerosa is (by definition) the name for the more widespread. For the less-widespread, perhaps the best course of action, only at this point, would have been to find a good-fitting junior synonym of L. praerosa to resurrect. Perhaps virgata (Lea 1841) is that synonym [14]. My nominee would be subglobosa (Say 1825). But in no case and at no time in this entire process are any additional newly-described pleurocerid nomina wanted on the floor of the Augean Stables [15]. I think Nathan and I are together on that.
Alas, none of the above happened in 2013. If I were 20 years younger, the study as I have outlined it would have taken place in 2014 – 2015, using allozyme markers, which are much more powerful than sequence data, but alas again, I am not. So, if any of you bright young students out there are shopping for a research project, I’ve got a great one all laid out for you on the banks of the Nolichucky. In fact, I’ve already got the manuscript written for you, like a malacological ChatGPT.
Go back and re-read the series of three essays I posted in the fall of 2016 on the cryptic Pleurocera of Maryville [16]. You have my permission to copy and paste all that text into a fresh manuscript, substitute Leptoxis for Pleurocera and Nolichucky River for Pistol Creek, submit to Science or Nature, and call a press conference. Your next big NSF grant is assured, your tenured professorship awaits.
Here's the bottom line. A critical reading of Nathan Whelan's (2013) dissertation research confirms the "periwinkle hypothesis" as I stated it last month, with an asterisk. Nathan's Baysian phylogeny, based on a concatenation of four genes, supports six biological species of Leptoxis: carinata, praerosa, crassa, picta, plicata, and arkansensis, with evidence of a seventh species, dubiously named virgata, cryptic under praerosa. The 6* Species Model for the North American pleurocerid genus Leptoxis is born.
Notes:
[1] In the original version of this essay, posted Tuesday afternoon 9May23, I stated that I first met Nathan in July of 2011 at the AMS. Nathan corrected me in an extensive comment posted shortly thereafter. His original 9May comment was subsequently deleted by Google, however, possibly because of some of the inflamatory language it included. So I resurrected his comment from the blogspot archives, emailed it back to Nathan, and he cleaned it up, and re-posted it on 11May. Look way down below.
[2] Whelan, Nathan V. (2013) Conservation, life history and systematics of Leptoxis Rafinesque 1819 (Gastropoda: Cerithioidea: Pleuroceridae). PhD Dissertation, University of Alabama, Tuscaloosa. 179 pp.
[3] Dillon, R T. and J. D. Robinson (2009) The snails the dinosaurs saw: Are the pleurocerid populations of the Older Appalachians a relict of the Paleozoic Era? Journal of the North American Benthological Society 28: 1 - 11. [pdf] To review the state of our knowledge at that (relatively early) stage in our understanding of the phenomenon we subsequently termed “mitochondrial superheterogeneity,” see:
- The snails the dinosaurs saw [16Mar09]
[4] Whelan, N.V. & E. E. Strong (2016) Morphology, molecules and taxonomy: extreme incongruence in pleurocerids (Gastropoda, Cerithiodea, Pleuroceridae). Zoologica Scripta 45: 62 – 87. To review the state of our understanding of mitochondrial superheterogeneity at this (much improved) state see:
- Mitochondrial superheterogeneity: What we know [15Mar16]
- Mitochondrial superheterogeneity: What if means [6Apr16]
- Mitochondrial superheterogeneity and speciation [3May16]
[5] There is some discrepancy between the number of branch tips depicted in Nathan’s Figure 5.4 and the number of samples as listed in his Table 5.2, possibly due to duplicate sequences. The sample sizes shown in my diagrammatic representation come from the Table, not the Figure.
[6] Here is Nathan Whelan’s (2013) Figure 5.4:
- Baysian phylogram inferred with the concatenated dataset with Cleopatra dropped for visualization purposes. [jpg] Numbers in front of nodes are posterior probabilities for each node. Scale bar meaningless.
[7] Whelan, N. V., P.D. Johnson, J.T. Garner, N.L. Garrison, & E.E. Strong (2022) Prodigious polyphyly in Pleuroceridae (Gastropoda: Cerithioidea). Bulletin of the SSB 1(2): 8419.
[8] I do have an hypothesis or two about the origin of “prodigious paraphyly,” however. Nathan’s Table 5.2, providing the sampling details for the 207 individual snails sequenced for his study, lists 63 populations, 31 of which had sample sizes of 1 – 2 and 32 had sample sizes 3 – 6. Of the latter 32, 13 demonstrated superheterogeneity ranging from 1 – 3. Given such a background sampling distribution, it seems likely to me that at least one or two populations of N = 5 (e.g., L. compacta) might be 100% superheterogeneous, pushing L. compacta (for example) entirely off the Leptoxis branches of Nathan’s tree.
[9] Dillon, R.T., and C. Lydeard (1998) Divergence among Mobile Basin populations of the pleurocerid snail genus, Leptoxis, estimated by allozyme electrophoresis. Malacologia. 39: 111-119. [pdf]
[10] Dillon, R. T., and S. A. Ahlstedt (1997) Verification of the specific status of the endangered Anthony's River Snail, Athearnia anthonyi, using allozyme electrophoresis. The Nautilus 110: 97 - 101. [pdf]
[11] The speciation of Leptoxis crassa from a Leptoxis praerosa population rendered L. praerosa paraphyletic. A cladist is any biologist who, upon first hearing the definition of the word, “paraphyly,” didn’t immediately react, “So, what?”
[12] Subspecies are populations of the same species in different geographic locations, with one or more distinguishing traits. For more, see:
[13] As fastidious as he is about the nodes of his gene trees, Nathan is cavalier with the branches. His Chapter 5 phylogenetic studies include no data on percent sequence divergence whatsoever. And (alas) I could not find any of his 2013 sequence data in GenBank. But the scale bar at the bottom of his Figure 5.4 is marked “0.4,” so eyeballing from that, it appears that the sequence divergence between his L. virgata cluster and his larger L. praerosa cluster is approximately 0.8 somethings.
[14] Isaac Lea described Melania virgata in 1841 from a single 0.3 inch (=7.6 mm) shell sent to him by Dr. Troost from “Tennessee.” Tryon demoted virgata to a “variety” of Anculosa subglobosa. But to my eye, and to that of Calvin Goodrich, Lea’s little figure of the shell, showing a relatively small aperture (“about half the length of the shell”) seems to depict a youngish Leptoxis carinata, a population of which has indeed jumped over the low hills from the New River into the Holston of Virginia, and could indeed have been picked up by Troost downstream in Tennessee. So Goodrich shifted virgata over to the genus Nitocris with carinata as he grouped Anculosa subglobosa together with A. praerosa. Burch followed Goodrich biologically, although not taxonomically, substituting Leptoxis for Anculosa, grouping virgata with carinata in the subgenus Mudalia, leaving praerosa in the typical subgenus. Bottom line is that in choosing to identify a putative species cryptic under praerosa as “Leptoxis virgata,” Nathan is following Tryon, and disagreeing with Goodrich, Burch, and Rob Dillon, all three of us together. Brash.
[15] This is an oblique reference to a very specialized, and hence very useful, reference work to the taxonomy of the Pleuroceridae:
- Graf, D. L. (2001) The cleansing of the Augean stables. Walkerana 12(27): 1 - 124.
[16] My three-part series on the rediscovery of Pleurocera gabbiana, cryptic under P. simplex in East Tennessee, was published both informally on this blog and more formally, in the FMCS Newsletter. See:
- [13Sept16] Dillon, R. T. (2016) Two reproductively isolated populations cryptic under Pleurocera simplex (Say, 1825) inhabiting Pistol Creek in Maryville, Tennessee. Ellipsaria 18(2): 15-16. [pdf]
- [14Oct16] Dillon, R. T. & J. D. Robinson (2016) The identity of the "fat simplex" population inhabiting Pistol Creek in Maryville, Tennessee. Ellipsaria 18(2): 16-18. [pdf]
- [14Nov16] Dillon, R. T. (2016) Match of Pleurocera gabbiana (Lea, 1862) to populations cryptic under P. simplex (Say, 1825). Ellipsaria 18(3): 10 - 12. [pdf]
Good to hear from you. So I really wanted to know the mean percent sequence divergence between your 7 populations of "L. virgata" and your 17 populations of L. praerosa. What is the conversion factor between "expected substitutions per site" and percent sequence divergence?
ReplyDeleteThere's not a direct conversion factor and I'm not going to revisit a dissertation from 9.5 years ago. It doesn't matter anyway. Based on what we know now, mtDNA should not be used for pleurocerid systematics. Neither should my dissertation. See my paper in BSSB.
ReplyDeleteNathan, I linked your paper last month. Sorry it's all come to this...
ReplyDeleteOK, I've got your BSSB paper open on my desk. Looking at the first paragraph of methods. You say, "Five species were sampled for transcriptome sequencing." Do you mean five individual snails? Or did you combine multiple individuals / multiple populations per species?
ReplyDeleteDude, it's all in there. I'm not going to hold your hand and walk you through the methods. There are tables with what we sampled, what individuals were sequenced and with what data generation approach was done for each individual. The transcriptomes were just for designing the probe set....
ReplyDeleteOMG. Rob, did you delete my comment? Did you not like being called out for being wrong?
ReplyDeleteYou dish it out on here all the time. Assuming you deleted it, I guess you couldn't take even a fraction of the criticisms you dish out.
Nathan - No, I did not delete your comment. I assumed that you did it. Weird. Let me look into the guts of the blogspot system and see if I can find it, and re-post it. Stand by.
ReplyDeleteNathan - I found your original comment of 5/9, but can't figure out how to restore it to the system. So I just copied it and emailed it to you. If you want to re-post, I'm standing by to approve.
ReplyDeleteThe comment that disappeared (with a small edit):
ReplyDeleteFor the record, nothing in my dissertation supports Rob's hypotheses. The explanation above is a gross mischaracterization of the data to fit preconceived notions and the idea that Rob has never been wrong about anything. The way the tree is collapsed has no biological/phylogenetic/systematic justification. It's just a visual slight of hand so Rob can falsely claim it supports his hypotheses.
Also I am not "...cavalier with the branches." Rob just doesn't seem to understand what the scale bar in Bayesian phylogenetics represent. They represent expected substitutions per site, not percent differences.
More importantly, my dissertation is outdated and not a good portrayal of the current state of the science. Why Rob chose to ignore my most recent phylogenetic study on Pleuroceridae is anyone's guess. Rob clearly knows about it because that's where the term "prodigious paraphyly" comes from (which, oddly, was not coined to explain patterns of pleurocerid mtDNA gene trees). My guess is that it'd be impossible to twist those data to support Rob's ideas. See here: https://ssbbulletin.org/article/view/8419. Then again, I'm sure Rob will find a way.
Also for the record, Rob first met me at FMCS in April 2011. I remember that so well because, in classic Dillion fashion, Rob tried to embarrass me (like he used to do after most student talks) after my talk. Instead, I had the whole room laughing at Rob's attempt to embarrass me because I pointed out how wrong it is to think pleurocerids haven't evolved since the Permian, as claimed in Dillon and Robinson (2009).
This note won't change Rob's mind, but hopefully others that have the misfortune of seeing this will understand how misguided the ideas in the post are.
I'll end with my reaction to the following quote from above. "...allozyme markers, which are much more powerful than sequence data":
HAHAHAHAHAHAHAHAHA. Bless your heart Rob.
What’s amazing is that this embarrassment of a blog post will appear in a future volume of FWGNA publication, without peer review and without inclusion of these comments which provide important context and rebuttal by researchers whose work is misrepresented, and it will be all yours for the low low price of $149.99!
ReplyDeleteYes, I am currently working on Volumes 5, 6, and 7 of the Freshwater Gastropods of North America series. The present essay will be featured in Volume 6, "Yankees at The Gap." No idea on the prices as yet. Stay tuned!
ReplyDelete