Thomas Nuttall (1786 – 1859) was a pioneering naturalist on
the American frontier, most famous as a botanist but with interests in geology,
ornithology, and yes, malacology as well.
In 1834 he resigned his professorship at Harvard and joined an
expedition up the newly-opening [1] Oregon Trail. Nuttall spent most of the next two years in
the Pacific Northwest, interrupted by an exursion to Hawaii, returning to a
position at the Academy of Natural Sciences of Philadelphia in 1836.
In 1841 our old buddy Isaac Lea [2] published a brief,
Latinate description of Lymnaea bulimoides [3], which he followed with an
English translation in 1844, as follows [4]:
"Shell ovately conical, rather thin, smooth, shining,
diaphanous, brownish yellow, slightly perforate; spire rather short; sutures
small; whorls five, slightly convex; aperture ovate. Hab. Oregon, Prof.
Nuttall."
Alas, Lea never published a figure of his Lymnaea
bulimoides. And the “Oregon” from which
Prof. Nuttall had just fetched that first sample was a vast territory that
included all of the modern states of Washington and Idaho, parts of Montana and
Wyoming and most of British Columbia [5].
By the blessings of Divine Providence, however, Lea’s type
lot has been preserved, even unto the present day. Both Haldeman [6] and Binney [7] published
little 1:1 figures of “authentic specimens,” as reproduced below. Prof. Haldeman [8] added a very similar
looking Limnea techella from Texas to the literature in 1867 as “surface
smoother than in L. bulimoides, of Oregon, with the lines of accretion less
apparent, and the labium more angular.”
His little 1:1 figure of L. techella is also reproduced below.
 |
| Hald. [6] Binney [7] Hald. [8] |
Now I feel quite
confident that a significant fraction of my (admittedly rather specialized)
readership will be at least passingly familiar with the crappy little lymnaeids
we find crawling around on the muddy margins of our rivers, ditches, and ponds
here in the American East. You all
listen up. None of you would ever
confuse a population of lymnaeids bearing shells such as those depicted above
with Lymnaea (Galba) cubensis/viator, am I right? The body whorl is way too big. And – good grief – look at the scale bar on
that holotype! Adult L. bulimoides often
reach double-digit shell lengths, whereas none of our crappy little Galba-type
lymnaeids on this side of the Mississippi River ever really do.
Nevertheless, in 1891 Henry Pilsbry became the first in a
long line of professional malacologists to confuse L. bulimoides with L.
cubensis, in a survey of the malacofauna of the Yucatan peninsula [9]. He began by synonymizing L. umbilicata C.B.
Adams 1840 under L. cubensis Pfeiffer 1839.
Then he wrote:
"The typical cubensis ranges at least as far west as the
Mississippi River and eastern Texas.
West and southwest of this it gives place to L. techella Hald., and L.
bulimoides Lea. The last form may be
considered a geographic race or subspecies of the cubensis. L. techella Hald. is nearly identical with
umbilicata."
Pilsbry corrected himself, however, in a survey of the
Mollusca of the southwestern states he published with J. H. Ferriss in 1906
[10]:
“Lymnaea techella was formerly considered by one of us to be
a synonym or race of L. cubensis Pfr, and L. bulimoides was treated as a
variety of the same species. They are
certainly very similar, but cubensis has a more triangular and less broadly
developed columellar expansion.”
Then going beyond a simple resurrection of Isaac Lea’s L.
bulimoides, Pilsbry and Ferriss went on to recognize three subspecies
underneath it: Haldeman’s techella from Texas, New Mexico and Arizona,
Hemphill’s sonomaensis from California, and their own new cockerelli,
widespread in New Mexico, Colorado, Nebraska and South Dakota.
Everybody look with me now at the three Pilsbry &
Ferriss figures I have reproduced below.
They’re all significantly larger than our crappy amphibious lymnaeids
here in The East, right? The shell
lengths reported by Pilsbry for all (N = 16) specimens he measured of all
subspecies ranged from 8 mm up to a whopping 14 mm, with mean = 10.2 mm, good
grief! Very, very clearly not L.
cubensis.
 |
| From Pilsbry & Ferriss [10] |
Pilsbry’s 1906 dabbling with the obscure little lymnaeids of
the American West did not take place in isolation, of course. Indeed, the flood of pulmonate gastropod
descriptions that washed across North America in the mid-nineteenth century
became a torrent in the early twentieth, our hero Frank Collins Baker surfing
high upon its crest. In 1909, Baker [11]
raised Hemphill’s sonomaensis to the full species level and described Lymnaea
hendersoni from Colorado, a new species “at first thought to be Lymnaea sonomaensis,”
but “differing in the form of the spire and aperture.”
And in 1911 he published his landmark “Lymnaeidae of
North America, Recent and Fossil [12],” placing his own contributions, and those of
his mentor Pilsbry [13], into a continental framework. Baker recognized four subspecies of Galba
bulimoides: the typical form restricted to the West Coast (BC, WA, OR, CA),
Haldeman’s techella ranging from California through the desert southwest to
Texas, Oklahoma, and Kansas, and Pilsbry’s cockerelli overlapping both, while
extending further north into Nebraska and The Dakotas. To these he added a new subspecies L.
bulimoides cassi from California, utterly indistinguishable from techella in all
respects, as well as the full species sonomaensis and hendersoni, both indistinguishable from cockerelli.
Although Baker carefully noted radula morphology when any
observations were available to him throughout his 1911 monograph, he did not
begin to draw a distinction between species bearing bicuspid first laterals and
tricuspid first laterals until 1928 [14].
He did note that the radula of G. bulimoides cockerelli bore bicuspid
first lateral teeth, “similar to that of cubensis” in 1911, and that
hendersoni also bore bicuspid first laterals “similar to those of techella,”
but offered no observations on any of the other taxa mentioned above, including
(oddly) techella.
There is no evidence that F.C. Baker ever confused L.
bulimoides, or any of the bulimoides-related taxa, with L. cubensis, or any
cubensis-related taxa, at any point in his illustrious career. He was keenly alert to even the finest
distinctions in phenotype, and ever ready to recognize new species and
subspecies on that basis. In 1919 he
described a Galba alberta from western Canada, to my eye looking like a dwarfed
elodes, with bicuspid first laterals [15].
In 1929 he teamed up with Junius Henderson to describe a Fossaria
perplexa from Washington state [16]. And
in 1939 he added a fresh subspecies Stagnicola [17] bulimoides vancouverensis,
distinguishing a strikingly large-bodied population from British Columbia [18].
 |
| From Leonard [19] Plate 1 |
In 1959 A. Byron
Leonard published a thorough and influential review of the entire gastropod
fauna of Kansas [19]. And I feel certain
that he must have had a copy of Baker’s 1911 monograph on his desk, showing the
range of both G. bulimoides techella and G. bulimoides cockerelli extending
through the Jayhawk State. In fact,
Baker listed five localities for techella in Kansas, although none for
cockerelli. Remember that. Baker also (of course) included Kansas within
the ranges of G. humilis and G. obrussa [20], both of which he considered
elements of the continental fauna broadly, but did not consider that the range
of G. cubensis extended as far north as Kansas.
So, Byron Leonard can be excused for identifying L.
bulimoides techella in Kansas, and not identifying L. cubensis. His Plate 1 is reproduced above, showing what
appears to be an unusually large [21] L. cubensis/viator shell identified as “L.
bulimoides techella.” This seems to be
a fresh re-emergence of the bulimoides/cubensis confusion independent of
Pilsbry’s 1891 error.
A third, independent confusion of bulimoides and cubensis
also has its roots in the soil of F.C. Baker but germinated much further
north. Baker provided neither figure nor
radular observations for the Fossaria perplexa he described with Junius
Henderson from Washington state in 1929 [16].
But his description (“resembles both parva and dalli … larger than dalli
and smaller than parva”) strongly suggests a synonym of either L. humilis or L.
cubensis/viator. In 1973, however, Arthur
Clarke [22] reported the discovery of a population of crappy little amphibious
lymnaeids in Alberta bearing shells “identical with type specimens of F.
perplexa” on their backs and radulas with bicuspid first laterals in their
mouths. Since he considered L. cubensis
“subtropical and tropical” in its distribution, Clarke reasoned that perplexa
must be “a hitherto unrecognized morph of the highly variable Lymnaea
bulimoides.”
And if the shell morphology of L. bulimoides is variable
enough to include a population that looks like L. perplexa, surely we might
also include populations that look like L. alberta, yes? Clarke did not have any original observations
to add in 1973, but on the basis of Baker’s original description of the radula
[15], lowered L. alberta to the status of “morph” under L. bulimoides as well
[23].
 |
| Clarke's [22] "morphs" of L. bulimoides |
With the advent of the 1980s came Jack Burch’s “North
American Freshwater Snails,” destined to enter the holy canon of American
malacology [24]. Burch recognized seven
subspecies of Fossaria (Bakerilymnaea) bulimoides: the three of Pilsbry
(bulimoides ss, techella, cockerelli), the two added by Clarke (alberta,
perplexa), the vancouverensis added by Baker, and Baker’s hendersoni, which had
heretofore been considered specifically distinct. Burch followed Baker in recognizing
sonomaensis at the species level, but clean forgot Baker's cassi, no big loss. Only techella, cockerelli, and the typical
subspecies were figured for bulimoides in the Burch Bible, plus sonomaensis as a separate species.
And so it came to pass that in January of 2022 I rendezvoused
with our good friend Bruce Stephen in Lawrence, KS, to review the extensive
freshwater gastropod holdings of the Kansas Biological Survey 1971 – 1981. You might remember Bruce from the
comprehensive survey of historic freshwater gastropod records from Nebraska
[25] he published back in 2015. Bruce
defended his dissertation, a modern survey of freshwater gastropods across
Nebraska and South Dakota, in 2018.
Bruce and I spent the week pulling vials of snails out of
metal cabinets on the fourth floor of Haworth Hall on the campus of the
University of Kansas, ultimately reviewing an impressive 642 lots, identifying
14 samples of Lymnaea humilis, 15 samples of L. cubensis/viator, and zero
samples L. bulimoides demonstrating the typical (or “techella”)
morphology. I feel confident that, sitting in these same precincts back in 1959, Byron
Leonard [19] confused L. cubensis/viator with L. bulimoides techella.
Indeed, Bruce has never confirmed a population of typical L.
bulimoides in Nebraska, or South Dakota, or North Dakota, for that matter. It would appear that the range of L.
bulimoides has been greatly exaggerated, almost certainly by confusion with L.
cubensis.
 |
| From Bruce's camera 1/22 |
Bruce and I did confirm 5 lymnaeid populations bearing
shells of the
cockerelli form in Kansas, with similar populations scattered
through Nebraska and The Dakotas as well.
Did F. C. Baker
[12] confuse
L. bulimoides techella with
L. bulimoides
cockerelli? We’ll come back to that
question next month.
But returning to the bulimoides/cubensis confusion, and
shifting one state south, to Oklahoma.
GenBank holds just two pair of sequences labeled “bulimoides:” a 16S/CO1
pair from E. A. Remigio [26, 27] and a 16S/CO1 pair from Wethington & Lydeard
[28]. The former pair (AF485657 and
AY227367, respectively), from an individual collected in “Oklahoma” (no further
information), are both 99% similar to the big body of sequence data for Galba
cubensis/viator that has accumulated in GenBank over many years.
The Remigio sequences were swept up into the 2011 study of
Correa et al. [29] and the influential 2021 study of Alda et al. [30],
prompting both of those sets of authors, and me myself a sinner [31], to
hypothesize that bulimoides might be a junior synonym of cubensis/viator in a
pair of posts on this very blog. Writing
here today, I feel quite certain that sequences AF485657 and AY227367 were
misidentified at their deposition. And I
have added red-font retractions to the bottoms of my blog posts of [7Aug12] and
[6July21].
The pair of 16S/CO1 sequences uploaded by Wethington &
Lydeard, EU038315 and EU038362 respectively, are 8.9% and 16.5% different from
the Remigio sequences, respectively, and hence did not get swept up into the
big worldwide surveys of Correa and Alda.
Blasting them against GenBank, however, both return close matches to
sequences obtained from a topotypic population of Lymnaea (Stagnicola) caperata,
deposited by Morningstar et al [32]: 98 – 99% for 16S and 96-97% for CO1. The only conclusion I think it is safe to
make at present from the negligible DNA data available for bulimoides is that I
am not going any further down this rabbit hole [33].
So let us now set the record straight, for all time. Lymnaea (Galba) bulimoides is a distinct,
valid biological species, not to be confused with Lymnaea (Galba)
cubensis/viator. Fossaria perplexa Baker
& Henderson 1929 is not a subspecies, synonym or morph of bulimoides, nor
is Galba alberta Baker 1919.
And in conclusion, Brothers and Sisters, I rise to the
pulpit. The confusion and
misunderstanding that has historically surrounded the crappy little amphibious
lymnaeids of western North America is but an extension of a greater darkness
that benights international malacology across five continents, Old World and
New. The figure below is from the 2011
review of neotropical lymnaeids published by Ana Correa and her colleagues [35],
as reproduced in my review of [7June21].
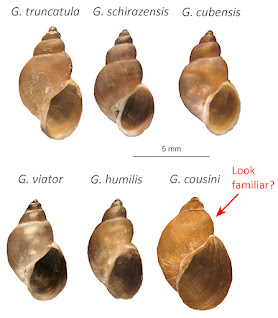 |
| From Correa et al. [33] |
Populations of crappy little amphibious lymnaeids identified
as “Galba cousini (Jousseaume, 1887)” are common and widespread in muddy
ditches and ponds on the Pacific side of South America, primarily in Ecuador
and Colombia. Where have you seen snails
bearing shells looking like that before?
All the lymnaeid populations we have discussed in this
overly long essay, and all of those depicted in Ana Correa’s figure above, are
potential hosts for the livestock fluke, Fasciola. In Central and South America, huge
international teams of malacologists and parasitologists have published
mountains of research on the evolutionary relationships among truncatula,
“schirazensis,” cubensis/viator and – yes – cousini. A quick search of GenBank returns 35 hits for
G. cousini alone.
Meanwhile here in the USA, the richest country on earth, the
leader of the free world, we have zero authentic sequences for any population
of our own Lymnaea (Galba) bulimoides, known to be an important host of
livestock fluke across the Pacific Northwest since 1929 [36]. We have four spurious mtDNA sequences from two crappy snails, both of which I think were
misidentified.
United States malacology had a two-generation head start on
South American malacology. Lea (1841)
trumps Jousseaume (1887) by 46 years. I
do not know how we have fallen so far behind the rest of the world today, but I
do know a continent-scale mess when I see it, and international embarrassment
when I feel it. Malacologists of America,
we must do better.
Notes:
[1] Although pioneered for foot traffic as early as 1811,
the Oregon Trail did not become passable by wagon until the 1830s.
[2] For a brief biography of “The Nestor of American
Naturalists,” see:
- Isaac Lea Drives Me Nuts [5Nov19]
[3] Lea, I (1841) On fresh water and land shells
(continued). Proceedings of the American
Philosophical Society 2(17): 30 – 34.
[4] Lea, I. (1844/46) Continuation of Mr. Lea’s paper on
fresh water and land shells.
Transactions of the American Philosophical Society 9(1): 1 – 31.
[5] The U.S. / Canadian boundary in the Pacific Northwest
was not established until 1846.
[6] Haldeman, S.S. (1844) A monograph of the freshwater
univalve Mollusca of the United States, Number 7 Philadelphia: Cary & Hart, Dobson, and
Pennington. 32 pp, 4 plates.
[7] Binney, W.G. (1865) Land and fresh water shells of North
America Part II, Pulmonata Limnophila and Thalassophila. Smithsonian
Miscellaneous Collections 143: 1 – 161.
[8] Haldeman, S.S. 1867
Description of a new species of Limnaea.
American Journal of Conchology 3: 194.
[9] Pilsbry, H.A. 1891 Land and Fresh-water mollusks
collected in Yucatan and Mexico.
Proceedings of the Academy of Natural Sciences of Philadelphia 43: 310 –
334.
[10] Pilsbry, H.A. and J.H. Ferriss (1906) Mollusca of the southwestern states II. Proceedings of the Academy of Natural
Sciences of Philadelphia 58: 123 – 175.
[11] Baker, F.C. (1909) A new species of Lymnaea. The Nautilus 22: 140 – 141.
[12] Baker, F.C. (1911) The Lymnaeidae of North and Middle
America, Recent and Fossil. Chicago
Academy of Sciences, Special Publication Number 3. 539 pp.
For a brief biography of our hero, see:
- The Legacy of Frank Collins Baker [20Nov06]
[13] For an exploration of the relationship between Frank
Collins Baker and Emperor Henry Augustus Pilsbry, see:
- The Emperor, the Non-child, and the Not-short Duct [9Feb21]
- Dr. Henry A. Pilsbry was a jackass [26Jan21]
[14] Baker, F.C. (1928) Freshwater Mollusca of Wisconsin,
Part I, Gastropoda. Bull. Wisc. Geol. Natur. Hist. Survey, no. 70. Madison:
University of Wisconsin Press. Baker
proposed Nasonia as a subgenus to distinguish species of Fossaria with bicuspid
lateral teeth, but alas, that name was preoccupied. The German malacologist W. K. Weyrauch
proposed the name “Bakerilymnaea” as a substitute in 1964.
[15] Baker, F.C. (1919) Fresh-water mollusca from Colorado
and Alberta. Bulletin of the American
Museum of Natural History 41(13): 527 – 539.
[16] Baker, F.C. and J. Henderson (1929) Fossaria perplexa
F. C. Baker and Junius Henderson.
Nautilus 42(3): 103-104.
[17] That's right, F. C. Baker himself transferred bulimoides from the genus Galba/Fossaria to the genus Stagnicola, simply because he discovered a population that was unusually large-bodied. There is absolutely no biological basis for recognizing genus (let alone subgenus) divisions in the worldwide Lymnaeidae. None. The FWGNA follows Hubendick in assigning essentially all lymnaeids to a single vanilla genus Lymnaea. We add subgenera for their indexing function only - just to help the Google machine find our research. For more, see:
- The Classification of the Lymnaeidae [28Dec06]
[18] Baker, F.C. (1939) Stagnicola bulimoides vancouverensis
nov. var. The Nautilus 52(4): 144.
[19] Leonard, A.B. (1959) Handbook of Gastropods in Kansas.
Miscellaneous Publications of the University of Kansas Museum of Natural
History 20: 1 – 224.
[20] Lymnaea (Galba) obrussa Say 1825 is a junior synonym of
Lymnaea humilis Say 1822. See:
- Exactly 3ish American Galba [6July21]
[21] The scale on Leonard’s entire Plate 1 is dubious. He stated, “figures enlarged approximately 2
times natural size,” but I do not know the original size of the printed
page. I’m working from a pdf.
[22] Clarke, A. (1973) The freshwater molluscs of the
Canadian Interior Basin. Malacologia, 13, 1-509
[23] The radula of Lymnaea (Stagnicola) elodes also bears
bicuspid first marginals. I do not agree
with Clarke about the synonymy of L. alberta, but am loathe to digress
further. It clearly is not
bulimoides. That's the point.
[24] This is a difficult work to cite. J. B. Burch's North American Freshwater
Snails was published in three different ways.
It was initially commissioned as an identification manual by the US EPA
and published by that agency in 1982. It
was also serially published in the journal Walkerana (1980, 1982, 1988) and
finally as stand-alone volume in 1989 (Malacological Publications, Hamburg,
MI).
[25] Stephen, B. J. (2015)
Species composition of Nebraska’s freshwater gastropod fauna: A review
of historical records. American
Malacological Bulletin 33: 61 – 71. For a review, see:
- Cornhusker Freshwater Gastropods [11May15]
[26] Remigio, E.A. and Hebert, P.D. (2003) Testing the utility
of partial COI sequences for phylogenetic estimates of gastropod
relationships. Mol. Phylogenet. Evol. 29
(3), 641-647.
[27] Remigio,E.A. (2002) Molecular phylogenetic relationships in
the aquatic snail genus Lymnaea, the intermediate host of the causative agent
of fascioliasis: insights from broader taxon sampling, Parasitol. Res. 88 (7),
687-696
[28] Wethington, A.R., & C. Lydeard (2007) A molecular
phylogeny of Physidae (Gastropoda: Basommatophora) based on mitochondrial DNA
sequences. Journal of Molluscan Studies
73: 241 - 257.
[29] Correa, A.C., J.S. Escobar, O. Noya, L.E. Velasquez, C.
Gonzalez-Ramirez, S. Hurtrez-Bousses & J-P. Pointier (2011) Morphological and molecular characterization
of Neotropic Lymnaeidae (Gastropoda: Lymnaeoidea), vectors of fasciolosis. Infection, Genetics and Evolution 11:
1978-1988. I reviewed that paper in my
post:
- The Lymnaeidae 2012: Fossarine Football [7Aug12]
[30] Alda, Pilar, M. Lounnas, A.Vázquez, R. Ayaqui, M.
Calvopiña, M. Celi-Erazo, R.T. Dillon Jr., L. González Ramírez, E. Loker, J. Muzzio-Aroca, A. Nárvaez, O.
Noya, A. Pereira, L. Robles, R. Rodríguez-Hidalgo, N. Uribe, P. David, P.
Jarne, J-P. Pointier, & S. Hurtrez-Boussès (2021) Systematics and
geographical distribution of Galba species, a group of cryptic and world-wide
freshwater snails. Molecular
Phylogenetics and Evolution 157: 107035. [pdf] [html] I reviewed that paper in my post:
- Exactly 3ish American Galba [6July21]
[31] I speculated that L. bulimoides might be a junior
synonym of L. cubensis/viator in both of the blog posts cited above. But in my own defense, see my footnote #11 of
6July21: “I am quite certain, however, that the single 16S sequence uploaded to
GenBank by Remigio, labeled “Fossaria bulimoides” but collected 2,000 miles
from the bulimoides type locality in Oregon, is weak evidence, indeed.”
[32] Morningstar,C.R., Inoue,K., Lang,B.K. and
Berg,D.J. (2018) A comprehensive status,
phylogenetic, and anatomical review of Stagnicola caperata (Say, 1829) in the
south-west United States. Aquatic Conservation 28 (3), 527-534.
[33] OK, maybe a little further. Our good friend Amy Wethington tells me that
she got her sequences from Rob Guralnick, who got them from a Mesa County,
Colorado sample identified as Lymnaea bulimoides by Shi-Kuei Wu. Shi-Kuei was a careful worker, and thumbing
through his (admirable) Colorado Inventory [34] I find no evidence that he was
confused about the identity of L. bulimoides.
I have no idea what happened here.
Classic GenBank SNAFU.
[34] Wu, S-K. (1989) Colorado Freshwater Mollusks. Natural
History Inventory of Colorado, no. 11. Boulder: Univ. Colorado Museum.
[35] Correa, A.C., J.S. Escobar, O. Noya, L.E. Velasquez, C.
Gonzalez-Ramirez, S. Hurtrez-Bousses & J-P. Pointier (2011) Morphological and molecular characterization
of Neotropic Lymnaeidae (Gastropoda: Lymnaeoidea), vectors of fasciolosis. Infection, Genetics and Evolution 11:
1978-1988.
[36] Shaw, J.N. and Simms, B.T. 1929. Galba bulimoides Lea
an intermediate host of Fasciola hepatica in Oregon. Science 69: 357.

























