Editor's Notes - This is the third installment of what will turn out to be a five-part series on the Lithasia of the Duck River in Middle Tennessee. It will help to familiarize yourself with my posts of [7Dec21] and [4Jan22] before continuing. This essay was subsequently published as: Dillon, R.T., Jr. (2023b) The third-most amazing research results ever published for the genetics of a freshwater gastropod population, and the fourth-most amazing, too. Pp 163 – 173 in The Freshwater Gastropods of North America Volume 6, Yankees at The Gap, and Other Essays. FWGNA Project, Charleston, SC.
In 2003, Russ Minton and Chuck Lydeard published a CO1 gene tree for the North American pleurocerid genus Lithasia in Molecular Ecology [2]. Nestled among the branches of that tree was the third-most amazing research result in the history of freshwater gastropod population genetics. What Minton and Lydeard found was… nothing.
The Minton & Lydeard study was very good by the standards of its day. Our colleagues did a thorough job collecting U1S2NMT3 individuals [7] from 30 Lithasia populations representing 11 nominal species and subspecies. We first touched on the M&L results back in [4Sept19], focusing on a single outside branch, labeled “L. geniculata pinguis.” And here is a quote from that 2019 essay: “To completely unpack the message being telegraphed to us by the enigmatic arboreal specimen (of Minton and Lydeard) would require at least 6 – 8 blog posts of standard length.” So, what follows is another installment [8].
Minton & Lydeard included in their analysis 19 Lithasia individuals from the main Duck River, identified as follows: 6 geniculata pinguis, 7 geniculata fuliginosa (from three sites), 1 geniculata geniculata, 4 duttoniana (from two sites), and 1 jayana. And on the basis of mtCO1 sequence, they were unable to distinguish among any of those 19 individuals. I was agog in 2003 and remain agog 20 years later.
 |
| Detail from Minton & Lydeard [2] fig 4, modified |
To contextualize. With no difficulty whatsoever, even at very small sample sizes, workers have easily been able to document 23% CO1 sequence divergence within Pleurocera simplex populations, 21% within Pleurocera catenaria, 19% within Pleurocera proxima, 15% within Leptoxis carinata, and 12% within L. ampla [9]. Then In 2003, in the pages of an international journal, Russ Minton and Chuck Lydeard stunned the world by reporting no more than a couple lousy nucleotides of difference in 19 individual Lithasia sampled down the 200-mile length of the Duck River, bearing five different Latinate nomina.
Well, maybe the M&L result is not terribly surprising for the 14 individual Duck River Lithasia geniculata they included in their survey, of the three subspecies. In December [7Dec21], we documented evidence of gene flow among subpopulations representing all three of those nomina, attenuated by distance but not much else [11]. The M&L pinguis sample seems to have been sampled from below the falls.
Nor am I terribly shocked by the absence of sequence divergence among the 4 duttoniana sampled by M&L. In January [4Jan22] we documented similar levels of isolation-by-distance between Duck River subpopulations bearing the DUT shell morphology that we had previously seen in the GEN form [12].
Nor am I even terribly surprised by the absence of sequence divergence between the 4 duttoniana and the singleton snail that M&L identified as “Lithasia jayana.” This is the first time the specific nomen “jayana” has appeared on the FWGNA blog. It will not be the last. We will have much more to say about Lithasia jayana in posts upcoming. But for now, please accept that there is no significant genetic difference between snails that have been identified as L. jayana on the basis of shell morphology and sympatric Lithasia populations that Goodrich, Minton, and everybody else has always called L. duttoniana.
Rather, the third-most amazing research result ever registered in the annals of freshwater gastropod population genetics is the absence of any detectable sequence divergence between the 14 individual geniculata (all subspecies) and the 5 individual duttoniana + jayana. Populations historically identified by those two sets of nomina really are morphologically distinct throughout their entire 100+ miles of sympatry in the Duck River, and everywhere else through their combined ranges across the Ohio, Cumberland, and Tennessee.
 |
| Can you tell us apart? [13] |
Isaac Lea and George Tryon and Calvin Goodrich and all modern workers even unto the present day have all drawn a clear and unambiguous distinction between Lithasia populations bearing a robust, oblong, bumpy shell morphology and those bearing a more acutely-spired shell morphology, with angular whorls often tuberculate or even slightly-spiny. In our January essay we abbreviated that former morphology “GEN” and that latter morphology “DUT.” Snails bearing shells of the DUT morphology range only up to around Duck River mile 186 (as opposed to 275 for GEN) and seem more common in the shallows, rather than on rocks in the middle. No prior worker has ever questioned the distinction between those two groups of taxa.
Setting 200 years of field observation aside, however. On the basis of their sequence data, Minton and Lydeard synonymized all the Duck River Lithasia taxa: pinguis, fuliginosa, duttoniana, and jayana, under Haldeman’s (1840) geniculata. Russ Minton then went on, in papers published in 2008 and again in 2018, to perform detailed morphometric analyses on the entire five-taxon GEN/DUT mishmash combined [14]. Bless his heart.
Well, the GEN and DUT populations do differ genetically, but not by much. When Johnson, Ahlstedt, and their colleagues sent me those Lithasia samples from Fountain Creek (site D), Wright Bend (site E) and Watered Hollow (site F) back in 2002, they divided them (quite naturally and conventionally) into oblong-bumpy subsamples they identified as Lithasia geniculata, and acute-angular subsamples they identified as Lithasia duttoniana. And in my January essay [4Jan22] I used the allozyme results I obtained from Wright Bend (site E) as an example of stable character phase disequilibrium between GEN and DUT. Results were the same at Fountain Creek and Watered Hollow. Hit this link for a pdf of my technical results [Ellipsaria 22(3)].
Lithasia bearing the GEN shell morphology and Lithasia bearing the DUT morphology sympatric in the Duck River do not constitute a single randomly-breeding population. There is some sort of reproductive isolation between them [15]. They are distinct biological species.
 |
| How many species can you see? Click to zoom [16]. |
But my goodness, the allozyme divergence between GEN and DUT is tiny! The gene frequencies I published in Table 1 of my paper in Ellipsaria 22(3) were certified in 2020 as the fourth-most amazing research results in the history of freshwater gastropod population genetics, at 88.7 international amazingness units [1].
Again, some context would seem to be in order. I ran allozyme gels on scores of pleurocerid populations during the 35 years I had access to a biochemical laboratory, 1980 – 2016. Typically, I would do an initial screening across 15 – 20 allozyme loci (17 in the case of the Duck River Lithasia), and then focus on the polymorphic loci for a detailed analysis. And very rarely did I ever find a pair of distinct biological species sharing alleles any more than at a couple loci, out of 15 or 20 [17].
Even among populations within pleurocerid species, fixed allozyme differences are not uncommon [18]. Across the 25 populations of P. proxima I surveyed for my 1984 dissertation, for example, it was possible to find conspecific populations sharing no alleles at five loci. And even within individual pleurocerid populations, sampled from single creeks or rivers, significant allozyme differences among subpopulations are not uncommon, as we witnessed in the P. proxima of Naked Creek in October [12Oct21], and in the L. geniculata of the Duck in December [7Dec21].
So seen in that context, to find no difference between a pair of reproductively-isolated pleurocerid species at 14 of 17 allozyme loci, and merely-statistical differences at the other three, shocked me back in 2002. And I’m obviously still not over it, any more than I am over the CO1 sequence results published by Minton and Lydeard in 2003. I was aghast at the time and remain aghast to this day.
Let this be a lesson to any of you high school seniors out there, looking for science fair projects. There are a lot of online purveyors of simple kits advertising “DNA barcoding” services, promising to identify any sort of unknown bug or slug you might pluck into a tube and mail to Canada. That’s fun, and I’m sure you’ll learn more from the experience than lying around your bedroom, watching Tik-Tok videos. But please understand that no serious scientist would ever publish a paper in the peer-reviewed literature relying on “DNA barcoding.”
Do I have time to touch on one additional feature of the 2003 M&L gene tree before you run out of patience with me this month? Notice this. Not only is there essentially zero divergence among their 19 Duck River samples of two reproductively-isolated species, we really don’t see much sequence divergence anywhere in the entire top half of the Minton & Lydeard Lithasia tree.
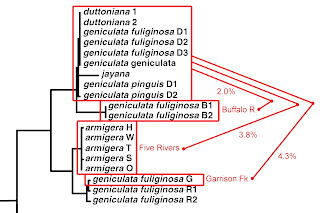 |
| Detail from Minton & Lydeard [2] fig 4, modified. |
If you back down one limb below the big Duck River cluster at the top, you’ll see a couple samples labeled, “geniculata fuliginosa” from 23 miles back up a tributary of the lower Duck River called the Buffalo. M&L did uncover 2.0% sequence divergence between their Duck River N = 19 and their Buffalo River N = 2, upon which basis Russ described a new species, “Lithasia bubala” in 2013 [19]. The allozyme data I reported on [7Dec21] did not support that [11].
Then if you back down two limbs from the M&L Duck River cluster, you find a set of five sequences identified as Lithasia armigera. These represent 14 individuals collected from five far-flung rivers: the Harpeth River and the Stones River (both tributaries of the Cumberland), the main Tennessee River way down in Alabama, the Wabash River (in Illinois) and the main Ohio River on the IL/KY border. All 14 of these snails, from five populations, were genetically indistinguishable. And all differed by just 3.8% from the Duck River group.
And if you back down three limbs from the M&L Duck River cluster, you’ll find a set of three sequences (representing 8 individuals) labelled “geniculata fuliginosa,” two from the Red River (a tributary of the Cumberland about 80 miles north of the Duck) and one sequence from Garrison Fork, an upstream tributary of the Duck River itself. The sequence divergence between that set of N = 8 and the set of N = 19 from the main Duck was 4.3%.
Let me say that again. There is less sequence divergence between L. duttoniana of the Duck River and L. armigera of the Wabash River almost 200 miles away, than between L. geniculata fuliginosa of the Duck River and L. geniculata fuliginosa of Garrison Fork, 25 miles upstream. What in the world does that mean? Stay tuned!
Notes
[1] I apologize for the overly-dramatic title. For the record, the CO1 sequence homogeneity in the Duck River Lithasia as reported by Minton & Lydeard in 2003 [2] scored 91.5 international amazingness units. The Bianchi et al. (1994) report of hybridization between P. virginica and P. semicarinata livescens [3] holds first place in the freshwater gastropod population genetics division at 93.2 international amazingness units, with Nathan Whelan’s [4] discovery of a wildebeest sequence in the population of bison he sampled at Shades Creek in second place at 91.9 iau.
For context, in the freshwater gastropod transmission genetics division, Yoichi Yusa’s discovery of multigenic sex determination in Pomacea [5] scored a whopping 98.7 iau in 2007, pushing Boycott’s (1923) paper on maternal inheritance of chirality in Lymnaea [6] to second all time, at 98.4 iau.
[2] Minton, R. L. and C. Lydeard. 2003. Phylogeny, taxonomy, genetics, and global heritage ranks of an imperiled, freshwater snail genus Lithasia (Pleuroceridae). Molecular Ecology 12:75-87.
[3] Bianchi, T. S., G. M. Davis, and D. Strayer 1994. An apparent hybrid zone between freshwater gastropod species Elimia livescens and E. virginica (Gastropoda: Pleuroceridae). Am. Malac. Bull. 11: 73 - 78.
[4] Whelan, N.V. & E. E. Strong (2016) Morphology, molecules and taxonomy: extreme incongruence in pleurocerids (Gastropoda, Cerithiodea, Pleuroceridae). Zoologica Scripta 45: 62 – 87. I reviewed Nathan’s findings in a series of posts back in 2016, see note [10] below.
[5] Yusa, Y. 2007. Nuclear sex-determining genes cause large sex-ratio variation in the apple snail Pomacea canaliculata. Genetics 175: 179-184. For more, see:
- Ampullariids star at Asilomar [11Aug05]
[6] Boycott, A.E. and C. Diver (1923) On the inheritance of sinistrality in Limnaea peregra. Proceedings of the Royal Society of London, Series B, Biological Sciences 95: 207 – 213.
[7] Usually 1, Sometimes 2, Never More Than 3. This has always been the rule-of-thumb in sampling for gene trees. See:
- The Lymnaeidae 2012: Stagnalis yardstick [4June12]
[8] Actually, looking back on this post from the bottom, I am afraid I have written an essay of twice what ought to be my standard length. And this is two installments. Sorry.
[9] I coined the term “mitochondrial superheterogeneity” on this blog in 2016 to describe double-digit intrapopulation sequence divergence [10]. Here are several prominent examples from the pleurocerids:
- Dillon, R. T., and R. C. Frankis. (2004) High levels of DNA sequence divergence in isolated populations of the freshwater snail, Goniobasis. American Malacological Bulletin 19: 69 - 77. [PDF]
- Lee, T., J. J. Kim, H. C. Hong, J. B. Burch, and D. O’Foighil (2006) Crossing the Continental divide: the Columbia drainages species Juga hemphilli is a cryptic member of the eastern North American genus Elimia. J. Moll. Stud. 72: 314-317.
- Dillon, R T. and J. D. Robinson (2009) The snails the dinosaurs saw: Are the pleurocerid populations of the Older Appalachians a relict of the Paleozoic Era? Journal of the North American Benthological Society 28: 1 - 11. [PDF]
- Dillon, R. T. Jr, and J. D. Robinson (2016) The hazards of DNA barcoding, as illustrated by the pleurocerid gastropods of East Tennessee. Ellipsaria 18: 22-24. [PDF]
- Whelan, N.V. & E. E. Strong (2016) Morphology, molecules and taxonomy: extreme incongruence in pleurocerids (Gastropoda, Cerithiodea, Pleuroceridae). Zoologica Scripta 45: 62 – 87.
[10] For more about the origin and significance of the phenomenon, see:
- Mitochondrial superheterogeneity: What we know [15Mar16]
- Mitochondrial superheterogeneity: What it means [6Apr16]
- Mitochondrial superheterogeneity and speciation [3May16]
[11] Dillon, R. T. (2020) Population genetic survey of Lithasia geniculata in the Duck River, Tennessee. Ellipsaria 22(2): 19 - 21. [PDF]
[12] Dillon, R. T. (2020) Reproductive isolation between Lithasia populations of the geniculata and duttoniana forms in the Duck River, Tennessee. Ellipsaria 22(3): 6 - 8. [PDF]
[13] From upper left: GEN, GEN, DUT, DUT, GEN, GEN.
[14] Papers in which Russ Minton lumped L. geniculata and L. duttoniana:
- Minton, R. L., A. P. Norwood & D. M. Hayes (2008) Quantifying phenotypic gradients in freshwater snails: a case study in Lithasia (Gastropoda: Pleuroceridae) Hydrobiologia 605: 173-182.
- Minton, R. L., K.C. Hart, R. Fiorillo, & C. Brown (2018) Correlates of snail shell variation along a unidirectional freshwater gradient in Lithasia geniculata (Haldeman 1840) (Caenogastropoda: Pleuroceridae) from the Duck River, Tennessee, USA. Folia Malacologia 26(2): 95 – 102.
[15] But I’ll bet dollars to donuts that they hybridize. I think hybridization is widespread in the North American family Pleuroceridae. See the paper by Bianchi et al from footnote [3] above.
[16] Five pleurocerid species are visible grazing across this rock in the Duck River at the Watered Hollow Boat Launch (RM 26): Pleurocera canaliculata canaliculata, Pleurocera laqueata laqueata, Leptoxis praerosa praerosa, Lithasia geniculata geniculata, and Lithasia armigera jayana. Notice that no juveniles are apparent whatsoever. All massively-shelled adults! I could write an entire essay on that phenomenon alone.
[17] A selection of papers showing typical levels of allozyme divergence between pleurocerid species:
- Dillon, R.T. and G.M. Davis (1980) The Goniobasis of southern Virginia and northwestern North Carolina: Genetic and shell morphometric relationships. Malacologia 20: 83-98. [PDF]
- Dillon, R. T., and S. A. Ahlstedt (1997) Verification of the specific status of the endangered Anthony's River Snail, Athearnia anthonyi, using allozyme electrophoresis. The Nautilus 110: 97 - 101. [PDF]
- Dillon, R. T. and A. J. Reed (2002) A survey of genetic variation at allozyme loci among Goniobasis populations inhabiting Atlantic drainages of the Carolinas. Malacologia 44: 23-31. [PDF]
[18] A selection of papers showing typical levels of allozyme divergence among populations within species:
- Dillon, R.T. (1984) Geographic distance, environmental difference, and divergence between isolated populations. Systematic Zoology 33:69-82. [PDF]
- Dillon, R.T. (1988) Evolution from transplants between genetically distinct populations of freshwater snails. Genetica 76: 111-119. [PDF]
- Dillon, R.T., and C. Lydeard (1998) Divergence among Mobile Basin populations of the pleurocerid snail genus, Leptoxis, estimated by allozyme electrophoresis. Malacologia. 39: 111-119. [PDF]
- Dillon, R. T. and J. D. Robinson (2011) The opposite of speciation: Population genetics of Pleurocera (Gastropoda: Pleuroceridae) in central Georgia. American Malacological Bulletin 29: 159-168. [PDF]
[19] Minton, R. L. 2013. A new species of Lithasia (Gastropoda: Pleuroceridae) from the Buffalo River, Tennessee, USA. The Nautilus 127:119-124.








" . . . no serious scientist would ever publish a paper in the peer-reviewed literature relying on “DNA barcoding.” Oh so brilliant!!!
ReplyDeleteThanks for your kind words,Old Buddy!
DeleteNo serious scientist would ever consider a non-peer reviewed screed in Ellipsaria a publication either.
DeleteGood to hear from you. And you make a good point. Science must be read critically, regardless of whether it is published in a glossy international journal or in a humble newsletter like Ellipsaria.
Delete